Understanding Genetic Variants and Disease Risk Using Over 100,000 Human Genomes


Anne O’Donnell-Luria, together with Gudmundsson and colleagues, investigated why some genetic variants thought to cause disease do not always result in illness. Using data from 807,162 individuals in the Genome Aggregation Database (gnomAD), they examined 734 “loss-of-function” variants in 77 genes linked to serious early-onset dominant diseases.
They found that 95% of these variants could be explained by biological factors such as low gene expression, alternative splicing, nearby compensating variants, or annotation errors. These mechanisms often prevent the variant from disabling the gene as predicted. Only 33 variants (4.5%) remained unexplained.
Their findings suggest that many variants previously considered pathogenic may actually be harmless, underscoring the need for caution when predicting disease risk in healthy individuals.
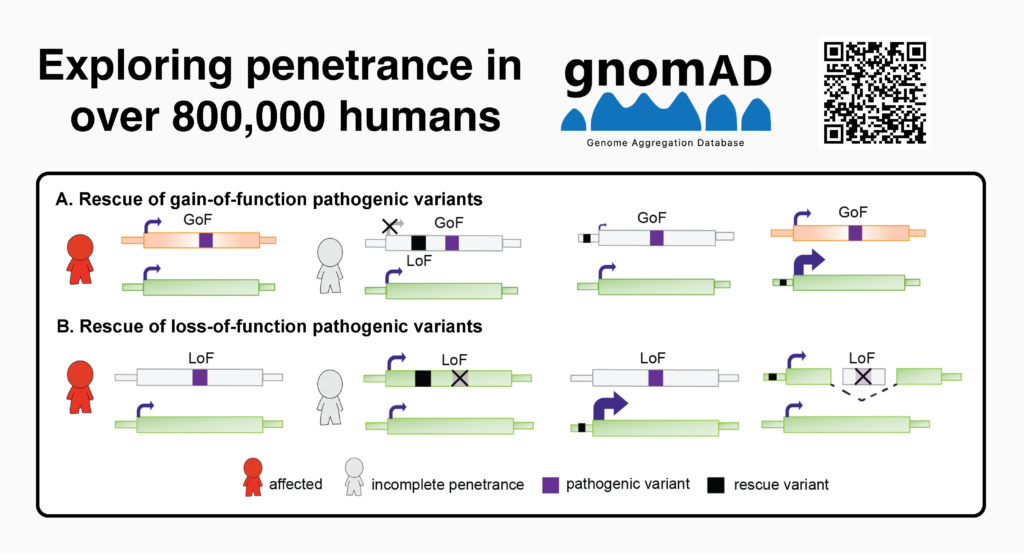
The study was published in October 2025: Exploring penetrance of clinically relevant variants in over 800,000 humans from the Genome Aggregation Database | Nature Communications.


